Why do I observe some ribosomal peaks after poly(A) selection?
Agilent's Bioanalyzer and other electrophoresis platforms often try to detect 18S and 28S rRNA peaks at all costs. Therefore, highly abundant transcripts are sometimes wrongly identified as rRNA.
In the traces below you will see that the Bioanalyzer software classifies two peaks within the trace of a poly(A)-selected sample as 18S and 28S rRNA (top figure). However, the overlay with diluted total RNA clearly demonstrates that those peaks are not rRNA peaks (bottom figure). Poly(A) selection was successful and no rRNA was left. This was also confirmed by RNA-Seq.
.png?inst-v=aefe7147-fd18-461a-93ce-29be806639e3)
Figure 1 | Bioanalyzer trace after the Poly(A) RNA Selection Kit was used with 5 µg of Universal Human Reference RNA (UHRR). The Bioanalyzer software wrongly classifies two peaks within the trace of the poly(A)-selected sample as 18S and 28S rRNA.
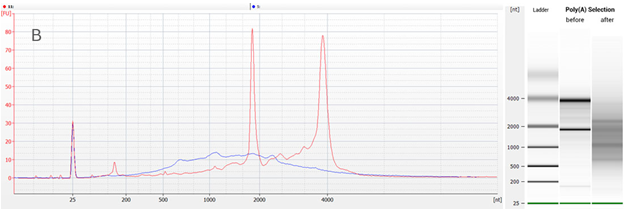
Figure 2 | Overlay of Bioanalyzer traces before and after the Poly(A) RNA Selection Kit was used. The overlay with diluted total RNA clearly demonstrates that those peaks identified as rRNA (top figure) are not rRNA peaks. Red trace: diluted total Universal Human Reference RNA (UHRR), intact (RIN 8.5). Blue trace: poly(A)-selected RNA.
