Are my libraries overcycled? If so, what should I do?
How do look like overcycled libraries?
A second peak between 1,000 – 9,000 bp, or an elevated baseline on a bioanalyzer trace that extends underneath the upper marker is an indication of overcycling (see Figure). Similar features can be seen with other types of microcapillary electrophoresis traces.
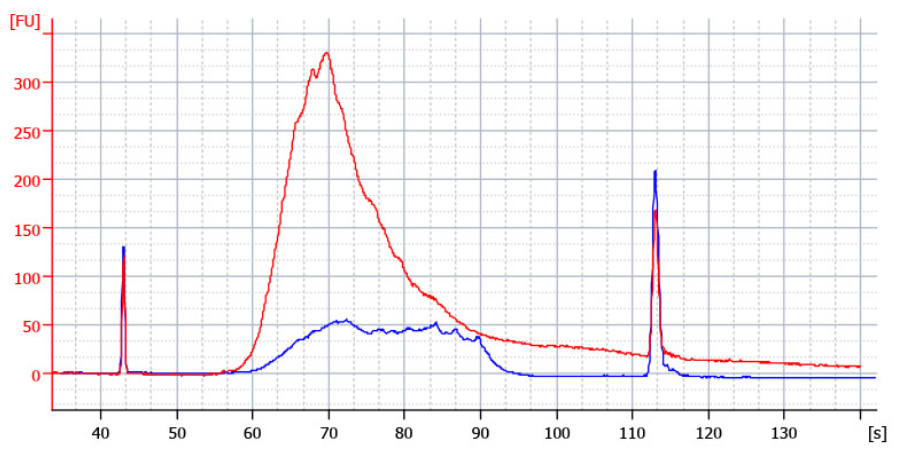
Figure | Correct versus overcycled QuantSeq Library Results: Correctly cycled (10ng UHRR, 19 cycles, blue), overcycled (10ng UHRR, 24 cycles, red). Low input protocol modifications were used (skipping step 2, 1 hour incubation at 42 degrees C at step 4, reduced volumes of PB and PS at steps 16 and 29).
What causes overcycling?
Overcycling occurs when too many PCR cycles are used for the final library amplification. In this case, the PCR reaction runs out of primers and template and generated ds-cDNA starts to denature and reanneal improperly. This results in longer bulky molecules that migrate at a lower speed on the Bioanalyzer chip or gels. This can interfere with exact library quantification for pooling and loading for sequencing if relying solely on the Bioanalyzer results.
How can I quantify overcycled libraries?
A qPCR assay (PCR Add-on kit, Cat. No. 208) for exact library quantification should be used in addition to Bioanalyzer or similar analyses, if such a high molecular weight peaks occurs and/or overcycling is suspected.
What is the effect of overcycling on my data?
Overcycled libraries can still be sequenced. However, these may have more PCR duplicates, which can affect quantification accuracy and sample clustering (i.e., by PCA). Overcycling may lead to a distortion in gene expression quantification and hence should be avoided where possible.
Should I sequence overcycled libraries or re-prep them?
In order to avoid possible biases in expression data, the best option would be to re-prep the libraries and use a lower number of PCR cycles for the library amplification. If the libraries cannot be re-prepped (i.e., due to precious or irreplaceable samples) it is best to proceed with sequencing.
How can I prevent overcycling?
Perform the qPCR assay to determine the optimal number of PCR cycles required for library generation.
For similar samples reduce the number of PCR cycles for library amplification (Endpoint PCR) to prevent over-cycling.
You can estimate the number of cycles to reduce according to the yield of overcycled libraries. To achieve half the yield, subtract 1 PCR cycle. To achieve 4x less yield, use 2 fewer cycles. To achieve 10x less yield, use 3 fewer PCR cycles.
