Do you have guidelines for the optimal S4U concentrations to use for different cell lines?
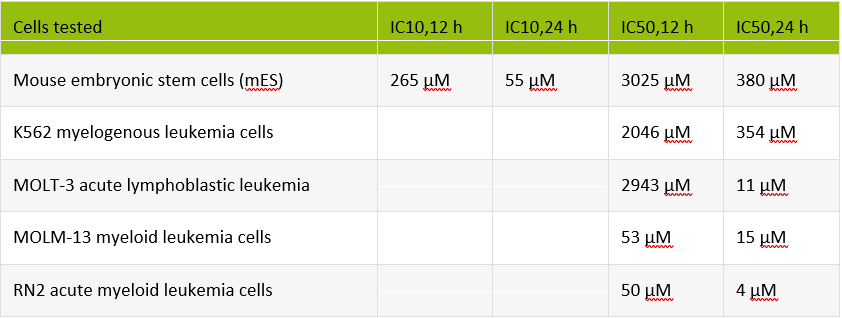
The following table provides S4U concentrations and IC10ti values (optimal labeling concentrations) that have been evaluated for various cell lines.
IMPORTANT: These concentrations are given as guidelines only; further optimization could be required. We recommend verifying optimal labeling concentrations using the SLAMseq Explorer Kit – Cell Viability Titration Module (Cat. No. 059) when establishing SLAMseq.
Additionally, as a starting point for planning your SLAMseq experiments we recommend a literature search to determine whether S4U concentrations for your specific cell type have been characterized before. Some references that have reviewed S4U concentrations are given below:
