Is QuantSeq suitable for preparation of libraries from degraded RNA or FFPE samples?
Yes!
Low quality and FFPE samples can be processed with our QuantSeq 3' mRNA-Seq library prep kits.
Our new QuantSeq FFPE 3' mRNA-Seq library prep kit (Cat. No. 222-223) has been specifically optimized for processing FFPE samples.
Our QuantSeq FWD and REV kits (Cat. No. 191-196 and 225) can also be used to prepare libraries from FFPE samples by following the protocol modifications for low quality samples outlined in the respective User Guides. For consistency reasons, we do not recommend switching to QuantSeq FFPE library prep kit if the newly generated data have to be compared with previous data obtained with the QuantSeq FWD or QuantSeq REV library prep kits.
The use of FFPE samples for QuantSeq-Pool is not recommended (please see this FAQ for more details).
Protocol Modifications
Some minor protocol modifications for QuantSeq FWD and REV are required and are indicated in the tables below:
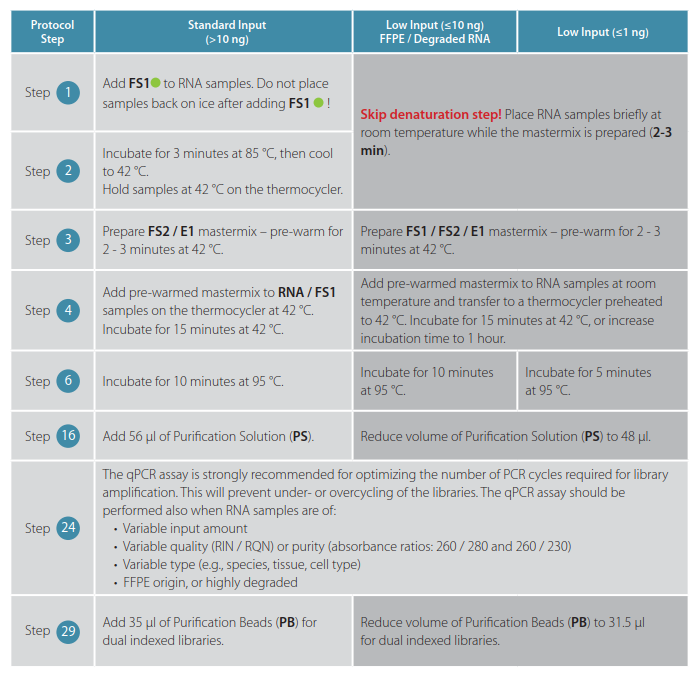
Table 1 | QuantSeq FWD V2 protocol modifications
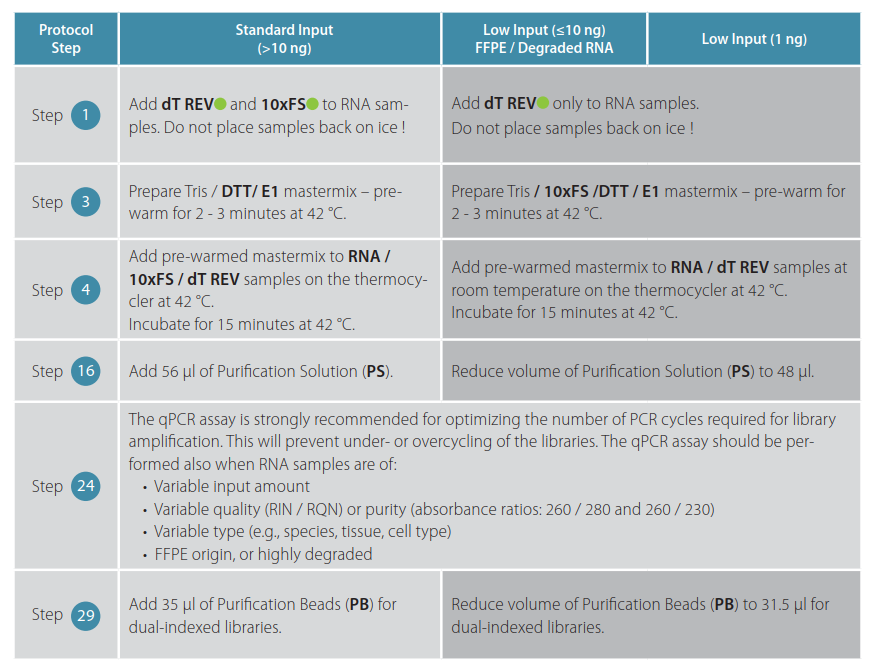
Table 2 | QuantSeq REV V2 protocol modifications
Preparing FFPE RNA for QuantSeq
The quality of RNA from FFPE tissues can vary greatly. We recommend measuring the DV200 value (the percentage of RNA greater than 200 nt in length) in addition to RIN values, as RIN values become less meaningful for highly degraded samples.
Optimizing PCR Cycle Numbers
Libraries prepared from FFPE RNA inputs are typically more variable, hence optimization of PCR cycle numbers is highly recommended. To prevent under- or over-cycling the libraries, we strongly recommend performing a qPCR assay to determine the optimal number of PCR cycles for the set of samples to be processed within each experiment.
When preparing libraries for comparative gene expression profiling from RNA samples that are of the same type, quality and input amounts, all libraries should be amplified using the same number of PCR cycles.
NOTE: The DV200 value is not a reliable predictor of the required number of PCR cycles.
If there is a range of optimal PCR cycle numbers predicted for your samples, you may wish to perform an additional endpoint PCR test to check the library yields are sufficient for sequencing. This is ideally done using half the library volume for a couple of samples that have different cycle number predictions (e.g., lowest, middle, and highest cycle number), and using the average number of cycles for the endpoint PCR (adding one additional cycle to account for using half the library volume). After purifying and quantifying these libraries, you can evaluate the relative yields and adjust the number of PCR cycles accordingly.
NOTE: Additional PCR and purification reagents provided in the PCR Add-on and Reamplification Kit V2 (Cat. No. 208) and the Purification Module with Magnetic Beads (Cat. No. 022.96) would be required for this type of endpoint PCR testing.
