Protocol for Whole Blood mRNA-Seq with Globin Depletion
Input amounts between 10 ng - 1 µg of total RNA can be used for poly(A) selection prior to CORALL RNA-Seq for Blood library preparation. RNA should be free of genomic DNA and other contaminants such as salts, metal ions, and organic solvents, which can be carried over from RNA extraction. Several sources of contamination can be detected with an UV-Vis spectrophotometer. An acceptably pure RNA sample should have an A260/A280 ratio between 1.8 and 2.1. The A260/A230 ratio should also be greater than1.8.
IMPORTANT: As blood samples contain ~10x more DNA than RNA, a DNase digest prior to RNA selection and library preparation is strongly recommended.
The RNAs should have an integrity score (RIN, RQN) of 7 or higher.
NOTE: The following protocol is a short version and first time users are advised to refer to the individual User Guides of Poly(A) RNA Selection, RiboCop Human/Mouse/Rat and CORALL RNA-Seq V2 for detailed protocols.
1. Poly(A) RNA Selection
Cat. No. 157 - Poly(A) RNA Selection Kit V1.5
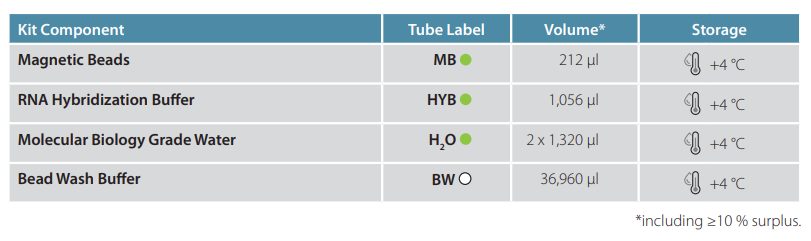
Aliquot and Wash Magnetic Beads
Transfer 2 µl of resuspended MB per reaction into a 1.5 mL tube. Place tube onto a magnet and discard supernatant.
Wash beads with 75 µl BW per reaction.
Discard supernatant and repeat wash step once for a total of 2 washes.
Discard supernatant and resuspend beads in 10 µl HYB per reaction.
Denature RNA
Prepare total RNA in a volume of 10 µl with RNase-free water.
Incubate for 1 min at 60 °C, then hold at 25 °C.
Hybridize Poly(A) RNA
Add denatured RNA (10 µl) to 10 µl of washed beads.
Incubate for 20 min at 25 °C / 1,250 rpm.
Wash 2x with 100 µl BW for 5 min at 25 °C / 1,250 rpm.
Withdraw and discard supernatant.
Elute Poly(A) RNA
Resuspend beads in 12 µl H2O.
Incubate for 1 min at 70 °C.
Immediately place onto a magnet for 5 min or until supernatant is clear.
Transfer the clear supernatant into a fresh tube. Safe stopping point.
Eluted Poly(A) RNA can be used as input for residual rRNA and globin depletion using RiboCop (Cat. No. 145, part of CORALL RNA-Seq for Blood, Cat. No. 185 - 186).
2. Residual rRNA and Globin mRNA Depletion
Cat. No. 145 RiboCop rRNA Depletion Kit for Human/Mouse/Rat plus Globin (HMR+Globin)
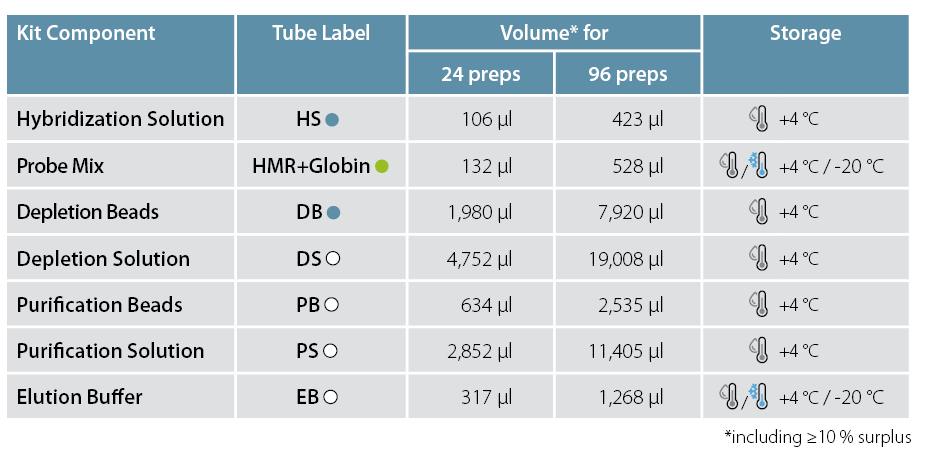
Hybridization
Prepare sample from step 14 in a total volume of 26 µl by adding H2O.
Add 4 µl HS.
Add 5 µl HMR+Globin and mix until homogeneous.
Denature for 5 min at 75 °C / 1,250 rpm.
Incubate for 30 min at 60 °C / 1,250 rpm.
Bead Washing
Resuspend DB, transfer 75 µl per reaction to a fresh tube.
Place onto a magnet for 2 - 5 min, discard supernatant.
Resuspend beads in 75 µl DS per reaction, incubate 2 min on magnet, discard supernatant.
Repeat Step 22 once for a total of 2 washes.
Resuspend beads in 30 µl DS per reaction.
Depletion
Spin down hybridized sample. Add 30 µl of prepared beads. Mix by pipetting 8x or until homogeneous.
Incubate for 15 min at 60 °C / 1,250 rpm. Spin down.
Place onto a magnet for 5 min.
Transfer 60 µl supernatant to a fresh tube and avoid disturbing the beads. ATTENTION: The supernatant contains the rRNA-depleted RNA.
Purification
Add 24 µl PB and 108 µl PS to supernatant, mix well, incubate for 5 min at RT.
Place onto a magnet for 5 - 10 min, discard supernatant.
Wash the beads twice with 120 - 150 µl 80 % EtOH, 30 sec. ATTENTION: Use 150 µl for 1.5 ml tubes.
Air dry beads for 5 - 10 min. ATTENTION: do not over dry the beads!
Add 12 µl EB, remove from magnet, mix well, incubate 2 min at RT.
Place onto a magnet for 2 - 5 min, transfer 10 µl of the supernatant to a fresh tube. Safe stopping point.
The RNA is now ready for library generation using CORALL RNA-Seq for Blood.
3. Whole Transcriptome Library Preparation
Cat. No. 171-175. CORALL RNA-Seq (CORALL RNA-Seq Library Prep is part of Cat. No. 185 and 186)
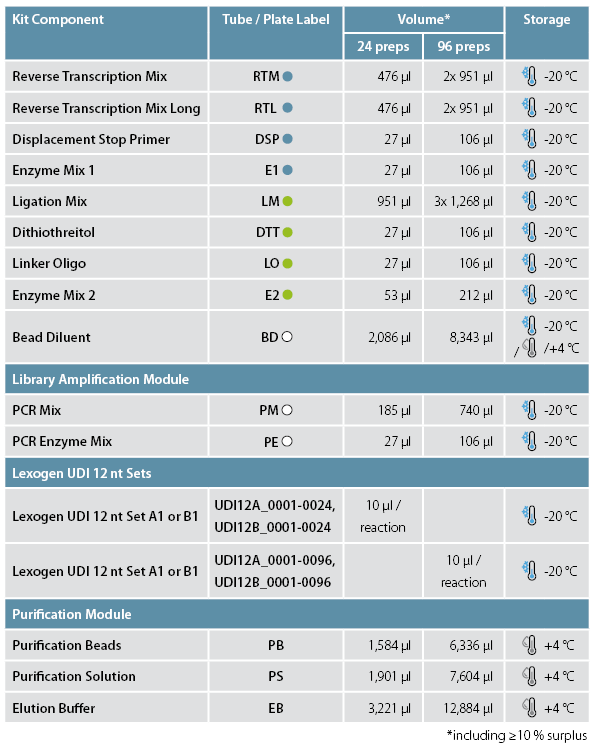
CORALL RNA-Seq libraries can be prepared with different insert sizes. For average library sizes of ~350 bp (~150 bp average insert size) follow the protocol for short insert sizes with RTM. For generation of libraries with an average size of ~550 bp (average insert size >150 bp) follow the protocol for long insert sizes with RTL.
3.1 Protocol for short insert sizes - RTM
Reverse Transcription
Prepare a mastermix of 14 µl RTM and 1 µl DSP per sample. Mix well.
Add 15 µl of RTM / DSP mastermix to 10 µl RNA sample. Mix well.
Incubate for 3 min at 94 °C, then 15 min at 16 °C.
Prepare a mastermix of 4 µl RTM and 1 µl E1 per sample. Mix well.
Add 5 µl RTM / E1 and mix well.
Incubate: 10 min at 25 °C, 40 min at 37 °C, 10 min at 42 °C, cool to 25 °C. Proceed immediately to purification!
Purification
Add 9 µl PB + 29 µl BD, mix well and incubate 5 min at RT.
Place onto a magnet for 2 - 5 min, discard supernatant.
Rinse beads twice with 120 µl 80 % EtOH, 30 sec.
Air dry beads for 5 - 10 min. ATTENTION: Do not let the beads dry too long!
Add 20 µl EB, remove from magnet, mix well, incubate 2 min at RT.
Place onto a magnet for 2 - 5 min, transfer the supernatant to a fresh PCR plate. Safe stopping point.
Linker Oligo Ligation
Prepare a mastermix of 36 µl LM, 1 µl DTT, 1 µl LO, and 2 µl E2 per sample. Mix well.
Add 40 µl of LM / DTT / LO / E2 mastermix to each sample. Mix well.
Incubate for 30 min at 37 °C, then cool to 25 °C. Proceed immediately to purification!
Purification
Add 9 µl PB + 50 µl BD, mix well and incubate 5 min at RT.
Place onto a magnet for 2 - 5 min, discard supernatant.
Add 30 µl EB, remove from magnet, mix well, incubate 2 min at RT.
Add 42 µl PS, mix well, incubate 5 min at RT.
Place onto a magnet for 2 - 5 min, discard supernatant.
Rinse beads twice with 120 µl 80 % EtOH, 30 sec.
Air dry beads for 5 - 10 min. ATTENTION: Do not let the beads dry too long!
Add 20 µl EB, remove from magnet, mix well, incubate 2 min at RT.
Place onto a magnet for 2 - 5 min, transfer 17 µl of the supernatant into a fresh PCR plate. Safe stopping point.
Optional: Perform qPCR assay to determine the optimal cycle number for library amplification. This step is strongly recommended for processing of blood RNA samples and requires the PCR Add-on and Reamplification Kit V2 (Cat. No. 208).
a. Add 2 µl of EB to the 17 µl of eluted cDNA.
b. Prepare a 2.5x stock of SYBR Green I nucleic acid stain (i.e., 1:4,000 dilution in DMSO; use Sigma-Aldrich, Cat. No. S9430 or ThermoFisher, Cat. No. S7585).
c. Combine 1.7 µl of cDNA with: 7 µl PM, 5 µl P5, 5 µl P7, 1 µl PE, 1.4 µl of 2.5x SYBR Green I nucleic acid stain, and 13.9 µl of EB, per reaction. Mix well.
d. PCR:
Step | PCR program | Cycles |
|---|---|---|
Initial Denaturation | 95 °C, 60 sec | |
Denaturation | 95 °C, 15 sec | 35x |
Annealing | 60 °C, 15 sec | |
Elongation | 72 °C, 60 sec | |
End Elongation | 72 °C, 6 min | |
Hold | 10 °C, ∞ |
e. Calculate the optimal cycle number for Endpoint PCR (please refer to 208UG591)
Endpoint PCR
Prepare a mastermix with 7 µl PCR Mix (PM) and 1 µl PCR Enzyme (PE) per reaction.
Add 8 µl of the PM / PE mastermix to 17 µl of the eluted library.
Add 10 µl of one Unique Dual Index Primer pair (UDI12A_0001-0384, or UDI12B_0001-0096) to each sample. ATTENTION: Reseal opened index wells after use! Use only one UDI / sample.
PCR:
Step | PCR program | Cycles |
|---|---|---|
Initial Denaturation | 95 °C, 60 sec | |
Denaturation | 95 °C, 15 sec | 11 - 25x according to qPCR result. |
Annealing | 60 °C, 15 sec | |
Elongation | 72 °C, 60 sec | |
End Elongation | 72 °C, 6 min | |
Hold | 10 °C, ∞ | Safe stopping point. |
Purification
Add 31.5 µl PB per reaction, mix well, incubate 5 min at RT.
Place onto a magnet for 2 - 5 min, discard supernatant.
Add 30 µl EB, remove from magnet, mix well, incubate 2 min at RT.
Add 30 µl PS, mix well, incubate 5 min at RT.
Place onto a magnet for 2 - 5 min, discard supernatant.
Rinse the beads twice with 120 µl 80 % EtOH, 30 sec.
Air dry beads for 5 - 10 minutes. ATTENTION: Do not let the beads dry too long!
Add 20 µl EB, remove from magnet, mix well, incubate 2 min at RT.
Place onto a magnet for 2 - 5 min, transfer 15 - 17 µl of the supernatant into a fresh PCR plate. Safe stopping point.
The libraries are now ready for QC and sequencing.
3.2 Protocol for long insert sizes - RTL
Reverse Transcription
Prepare a mastermix of 14 µl RTL and 1 µl DSP per sample. Mix well.
Add 15 µl of RTL / DSP mastermix to 10 µl RNA sample. Mix well.
Incubate for 1 min at 94 °C, then 15 min at 16 °C.
Prepare a mastermix of 4 µl RTL and 1 µl E1 per sample. Mix well.
Add 5 µl RTL / E1 and mix well.
Incubate: 10 min at 25 °C, 40 min at 37 °C, 10 min at 42 °C, cool to 25 °C. Proceed immediately to purification!
Purification
Add 9 µl PB + 6 µl PS, mix well and incubate 5 min at RT.
Place onto a magnet for 2 - 5 min, discard supernatant.
Rinse beads twice with 120 µl 80 % EtOH, 30 sec.
Air dry beads for 5 - 10 min. ATTENTION: Do not let the beads dry too long!
Add 20 µl EB, remove from magnet, mix well, incubate 2 min at RT.
Place onto a magnet for 2 - 5 min, transfer the supernatant to a fresh PCR plate. Safe stopping point.
Linker Oligo Ligation
Prepare a mastermix of 36 µl LM, 1 µl DTT, 1 µl LO, and 2 µl E2 per sample. Mix well.
Add 40 µl of LM / DTT / LO / E2 mastermix to each sample. Mix well.
Incubate for 30 min at 37 °C, then cool to 25 °C. Proceed immediately to purification!
Purification
Add 9 µl PB + 76 µl BD, mix well and incubate 5 min at RT.
Place onto a magnet for 2 - 5 min, discard supernatant.
Add 30 µl EB, remove from magnet, mix well, incubate 2 min at RT.
Add 42 µl PS, mix well, incubate 5 min at RT.
Place onto a magnet for 2 - 5 min, discard supernatant.
Rinse beads twice with 120 µl 80 % EtOH, 30 sec.
Air dry beads for 5 - 10 min. ATTENTION: Do not let the beads dry too long!
Add 20 µl EB, remove from magnet, mix well, incubate 2 min at RT.
Place onto a magnet for 2 - 5 min, transfer 17 µl of the supernatant into a fresh PCR plate. Safe stopping point.
Optional: Perform qPCR assay to determine the optimal cycle number for library amplification. This step is strongly recommended for processing of blood RNA samples and requires the PCR Add-on and Reamplification Kit V2 (Cat. No. 208).
a. Add 2 µl of EB to the 17 µl of eluted cDNA.
b. Prepare a 2.5x stock of SYBR Green I nucleic acid stain (i.e., 1:4,000 dilution in DMSO; use Sigma-Aldrich, Cat. No. S9430 or ThermoFisher, Cat. No. S7585).
c. Combine 1.7 µl of cDNA with: 7 µl PM, 5 µl P5, 5 µl P7, 1 µl PE, 1.4 µl of 2.5x SYBR Green I nucleic acid stain, and 13.9 µl of EB, per reaction. Mix well.
d. PCR:
Step | PCR program | Cycles |
|---|---|---|
Initial Denaturation | 95 °C, 60 sec | |
Denaturation | 95 °C, 15 sec | 35x |
Annealing | 60 °C, 15 sec | |
Elongation | 72 °C, 60 sec | |
End Elongation | 72 °C, 6 min | |
Hold | 10 °C, ∞ |
e. Calculate the optimal cycle number for Endpoint PCR (please refer to 208UG591)
Endpoint PCR
Prepare a mastermix with 7 µl PCR Mix (PM) and 1 µl PCR Enzyme (PE) per reaction.
Add 8 µl of the PM / PE mastermix to 17 µl of the eluted library.
Add 10 µl of one Unique Dual Index Primer pair (UDI12A_0001-0384, or UDI12B_0001-0096) to each sample. ATTENTION: Reseal opened index wells after use! Use only one UDI / sample.
PCR:
Step | PCR program | Cycles |
|---|---|---|
Initial Denaturation | 95 °C, 60 sec | |
Denaturation | 95 °C, 15 sec | 11 - 25x according to qPCR result. |
Annealing | 60 °C, 15 sec | |
Elongation | 72 °C, 60 sec | |
End Elongation | 72 °C, 6 min | |
Hold | 10 °C, ∞ | Safe stopping point. |
Purification
Add 31.5 µl PB per reaction, mix well, incubate 5 min at RT.
Place onto a magnet for 2 - 5 min, discard supernatant.
Add 30 µl EB, remove from magnet, mix well, incubate 2 min at RT.
Add 30 µl PS, mix well, incubate 5 min at RT.
Place onto a magnet for 2 - 5 min, discard supernatant.
Rinse the beads twice with 120 µl 80 % EtOH, 30 sec.
Air dry beads for 5 - 10 minutes. ATTENTION: Do not let the beads dry too long!
Add 20 µl EB, remove from magnet, mix well, incubate 2 min at RT.
Place onto a magnet for 2 - 5 min, transfer 15 - 17 µl of the supernatant into a fresh PCR plate. Safe stopping point.
The libraries are now ready for QC and sequencing.
