What are the differences between QuantSeq REV and FWD?
With QuantSeq Reverse (REV, Cat. No. 225), the Read 1 adapter sequence is located on the 5' end of the oligo(dT) primer and the Read 2 adapter sequence is located on the 5' end of the random primers used during second strand synthesis. This generates libraries that have a reverse orientation compared to those generated by the QuantSeq FWD kit, i.e., Reads 1 reflect the cDNA sequence and Reads 2 reflect the mRNA.
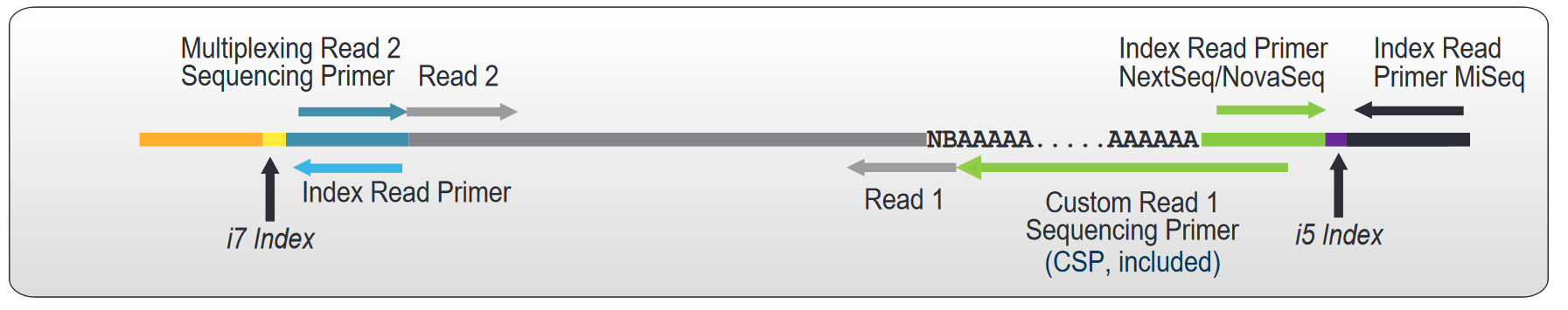
Figure | Schematic overview of the QuantSeq REV V2 library preparation workflow (Cat. No. 225). Sequencing read orientation for QuantSeq REV is depicted, with Read 1 (green) and Read 2 (blue).
A Custom Sequencing Primer (CSP, included in the kit) is required for sequencing QuantSeq REV libraries, to avoid sequencing the poly(T) stretch at the start of Read 1. This enables Read 1 to begin at the exact 3' end of the transcript.
Using Paired-End sequencing is advantageous to identify 3' UTR isoforms.
The use of QuantSeq REV is therefore ideal for applications including alternative polyadenylation site identification and alternative 3' UTR expression studies.
