What is the difference between QuantSeq FWD and QuantSeq REV protocols?
Both kit versions yield sequences close to the 3' end of transcripts. The difference is in the location of the Read 1 linker sequence. If it is located in the 5' part of the second strand synthesis primer (QuantSeq FWD V2, Cat. No. 191 - 196), NGS reads will be generated towards the poly(A) tail.
QuantSeq FWD is therefore recommended for all gene expression applications.
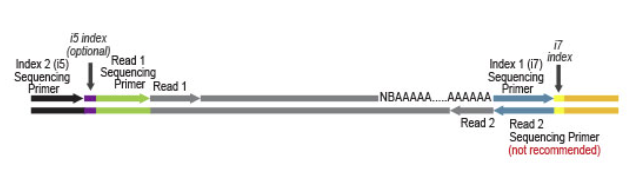
Figure 1 | Read orientation for QuantSeq FWD V2 (Cat. No. 191 - 196)
With QuantSeq Reverse (QuantSeq REV V2, Cat. No. 225), the Read 1 linker sequence is located on the 5' end of the oligo(dT) primer. A Custom Sequencing Primer (CSP, included in the kit) is required for sequencing in order to start the read directly at the 3' end. Based on this feature, the exact transcription end site can be determined.
QuantSeq REV is ideal for applications including alternative polyadenylation site identification and alternative 3' UTR expression studies.

Figure 2 | Read orientation for QuantSeq REV V2 (Cat. No. 225)
