What is the prominent peak around 350bp in my RTL CORALL V2 libraries generated from rRNA-depleted input RNA?
CORALL libraries generated with the RTL chemistry for longer insert sizes efficiently capture highly structured non-coding RNAs of ~300 nucleotides in size which are present in ribo-depleted RNA but not in poly(A) selected RNA. Examples include snoRNAs, snRNAs, or the RNA component of signal recognition particles (e.g., 7SL2).
CORALL library prep protocols use a fragmentation-free workflow, therefore the structure of these RNA species may lead to defined products in the final library. Inserts derived from these RNA species may therefore appear as distinct peaks in the library trace. Common RNA extraction and ribo-depletion protocols, such as RiboCop preserve RNA species < 300 nucleotides.
When present in the input RNA (Fig. A), peaks in the library trace directly correspond to these non-coding RNAs (Fig. B). The figures below show representative RNA and library traces generated from human reference RNA (UHRR) following ribo-depletion. Sequencing data quality is not affected by the distinct appearance of these RNAs in the libraries.
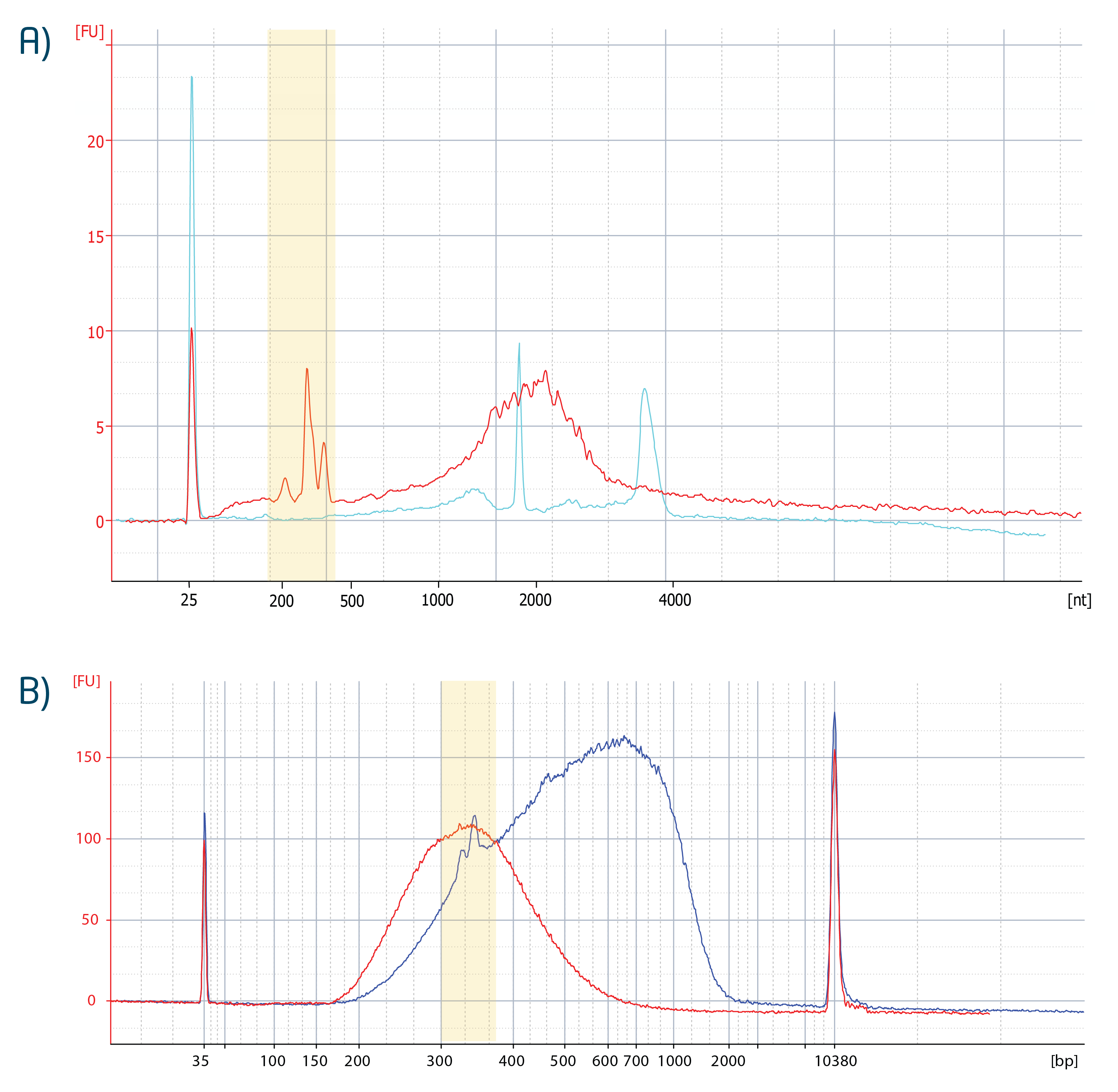
Figure | Bioanalyzer trace of ribo-depleted RNA and typical CORALL libraries generated from rRNA-depleted input RNA. Panel A: 500 ng Universal Human Reference RNA (UHRR) was used with Lexogen's RiboCop HMR V2 Kit and the resulting ribo-depleted RNA (red trace) is shown as overlay with 500 pg total RNA (cyan trace). Panel B: One fifth of the ribo-depleted RNA (corresponding to 100 ng total RNA) was inserted into CORALL. Either RTM (red trace) or RTL (blue trace) was used with the respective library preparation protocol. Purified libraries were quality controlled using a Bioanalyzer High Sensitivity DNA assay. Libraries were amplified with 12 PCR cycles. The RTM synthesized library (red trace) has an average library size of 355 bp. The RTL synthesizied library (blue trace) has an average library size of 593 bp, according to Bioanalyzer.
NOTE: The appearance of this peak in a library is dependent on the RNA input (e.g., species and presence / abundance of small RNA species).
