What is the recommended sequencing format and read depth?
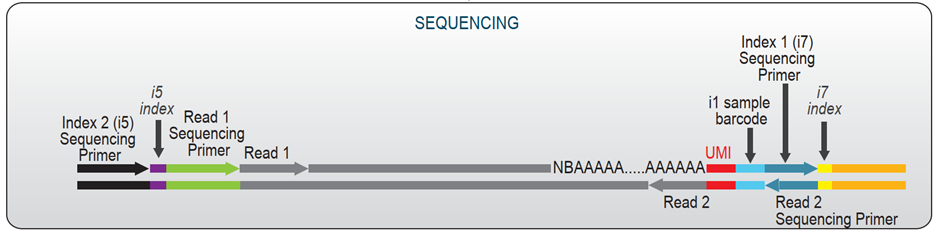
LUTHOR HD Pool libraries require asymmetric paired-end sequencing, as Read 2 contains the i1 sample barcode and Unique Molecular Identifier (UMI) sequence.
Read 1: Optimal read length is 75 - 100 bp.
Read lengths <50 bp are not recommended as shorter reads are more likely to be lost through trimming, resulting in lower alignment rates.
Read lengths >100bp can also be used. However, it is important to note that per base sequencing quality will typically decrease as read length increases, as more inserts will be fully covered and start to read into the poly(A) stretch and adapter sequences.
Regardless of read length, trimming of Read 1 should be performed as a first step in data analysis, to remove poly(A) homopolymers, adapter sequences and low quality bases. This will ensure you retain maximal insert read lengths for mapping purposes.
Read 2: Recommended read length is 12 - 24 bp to read-out the 12 nt i1 sample barcode (required) and 12 nt UMI (optional).
Using a read length of 24 bp for Read 2 is highly recommended in order to fully read-out the UMI to avoid gene quantification biases.
Longer read lengths are not recommended as this will capture the poly(T) stretch. Reading through homopolymer stretches can negatively impact read quality and will require more complicated trimming.
For sequencing depth, we recommend 1-5 M reads/sample. If you are mainly interested in highly expressed genes, a lower coverage can be considered.
