What level of depletion can be expected from RiboCop for Fish?
Processing 1 µg of high-quality Danio rerio RNA with RiboCop for Fish typically results in ~2% remaining rRNA reads, while processing 100 ng results in ~3,3% remaining rRNA reads (see figure below).
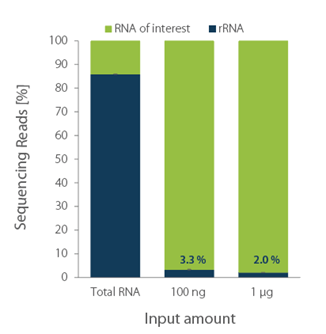
Figure 1. RiboCop rRNA Depletion Kit for Fish efficiently removes rRNA across a wide range of input amounts. NGS libraries were prepared using Lexogen’s CORALL RNA-Seq V2 Library Prep Kit. Successful depletion was monitored by NGS sequencing and subsequent analysis of remaining rRNA reads from 10 ng untreated (Total RNA) and depleted D. rerio RNA (100 ng and 1 µg). The percentage of reads mapping to rRNA is plotted in blue.
Moreover, RiboCop for Fish has been reported to deplete additional species, such as Gasterosteus aculeatus (reduction to ~1.4 % rRNA reads for 100 ng input RNA) and Perca fluviatilis (reduction to ~1.7 % rRNA reads for 100 ng input RNA).
Depletion rates may vary depending on the used species, quality of the RNA, purity of the RNA and the input amounts. Contaminants, such as salts, metal ions, and organic solvents, may have a negative impact on the efficiency of the protocol.
NOTE: The final observed percentage of rRNA reads after depletion can also vary depending on the quality of used reference genome.
