What is the CSP and how do I use it?
In QuantSeq REV libraries, Read 1 sequencing starts at the beginning of the poly(A) tail, reading back into the transcript sequence. Therefore a Custom Sequencing Primer (CSP Version 5, included in the kit) is required to avoid sequencing the poly(T) stretch at the start of Read 1 (See Figure below).
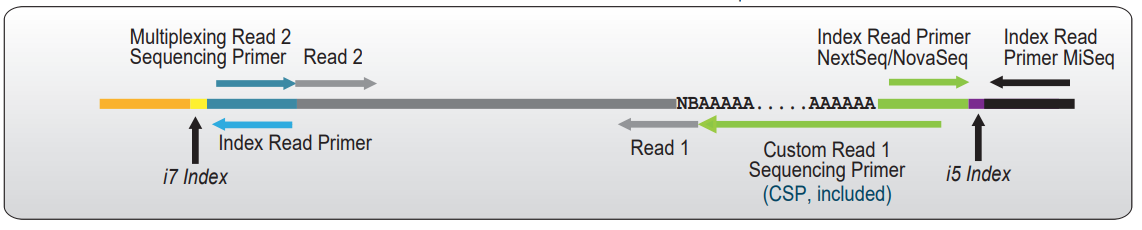
Figure: Sequencing Read orientation of QuantSeq REV libraries. Reads 1 start at the beginning of the polyA tail and reflect the cDNA sequence. The CSP contains a poly(T) stretch to cover the polyA tail and avoid quality drops and registration errors during the sequencing.
QuantSeq REV libraries can be sequenced on Illumina and Aviti sequencing platforms (please see this FAQ for more information on instruments supporting the use of Custom Primers). Preparation of the CSP may vary depending on the Sequencer/method used. Please find below some information on how to use the CSP on Illumina and Aviti instruments. Please keep in mind these guidelines may be subject to change should the sequencing reagents and/or platforms be updated.
Illumina platforms
The provided CSP replaces the Multiplex Read 1 Sequencing primer normally used for sequencing.
General guidelines to sequence your QuantSeq REV libraries on Illumina instruments:
QuantSeq REV libraries should not be sequenced together with libraries prepared with any other different kit. This is due to the use of the CSP Version 5, which will not prime other library types correctly.
PhiX spike-in should not be used for QuantSeq REV runs as it will not be primed by the CSP Version 5.
If loading amount guidelines are not provided, please use the upper range of loading amounts specified by Illumina for the instrument type.
The CSP should never be mixed together with the standard Illumina Read 1 Sequencing primer. A primer mixture would result in low clusters calls and the resulting reads would be contaminated by poly(T) stretches.
NOTE: We recommend that you also consult the relevant Custom Sequencing Primer Guides from Illumina and/or the respective Illumina Custom Primer Guide of the sequencing platform used and follow those instructions when preparing your CSP for use. If you have any questions regarding CSP use please contact our technical support team via support@lexogen.com.
ATTENTION: If you are sequencing your QuantSeq REV libraries with a service provider or sequencing facility, please provide the above relevant information to the facility, ALONG WITH THE CSP Version 5!
AVITI Platforms
To sequence your QuantSeq REV libraries on AVITI instruments, you can follow:
The spike-in method where diluted custom primers are spiked in the Cloudbreak Freestyle or Adept primer set tubes.
The replacement workflow where primers in the cartridge are replaced by buffer tubes (add the custom primers) from the Custom Primer Set Cloudbreak Freestyle or Adept Custom Primer Set Cloudbreak.
Detailed information can be find in the Cloudbreak Sequencing User Guide (MA-0058).
NOTE: When QuantSeq REV libraries are mixed with other Lexogen libraries, and they represent less than 25% of the total lanemix, the CSP is not required! In this case, we recommend adding 2% PhiX in the lanemix for sequencing (please see this FAQ for additional information).
IMPORTANT: Please note that for sequencing QuantSeq REV libraries on AVITI with CSP:
The provided CSP in QuantSeq REV V2 24 rxn kit (Cat. No. 225.24) is not sufficient for a sequencing run. Please add Cat. No. 245.01 to your order.
The provided CSP in QuantSeq REV V2 96 rxn kit (Cat. No. 225.96) is sufficient for a single sequencing run. If you plan to perform multiple runs, please add Cat. No. 245.01 to your order.
If you have more questions, please contact support@lexogen.com
