Can I use sample barcodes for multiplexing TeloPrime cDNA for NGS applications?
Sample barcoding can be included in two ways, either during reverse transcription through the use of custom reverse transcription primers (see Appendix D of the TeloPrime V2 User Guide), or by adding sample barcodes after full-length cDNA amplification is complete. Sample multiplexing is supported for long-read sequencing platforms as well:
PacBio IsoSeq
Sample barcodes can be included in alternative oligodT-primers and used in place of the standard Reverse Transcription Primer (RTP) for the first strand cDNA synthesis (steps 1 – 3). We recommend using the sample barcoded RT primer designs provided by PacBio for IsoSeq libraries (https://www.pacb.com/wp-content/uploads/Procedure-Checklist-Iso-Seq-Express-Template-Preparation-for-Sequel-and-Sequel-II-Systems.pdf).
These primers can be ordered as desalted oligos. The concentration of these oligos in the reverse transcription reaction may need to be optimized within the range of: 12.5 nM – 1.5 uM.
In addition, a custom RP primer will be needed for the Endpoint PCR, to use together with the standard FP primer. This custom RP primer needs to bind the 5' end of the barcoded RT primers and should have a melting temperature (Tm) matching, or as close as possible to that of the standard FP primer included in the TeloPrime V2 kit.
Oxford Nanopore
Sample barcodes can be introduced onto TeloPrime cDNA after endpoint PCR using the Oxford Nanopore PCR Barcoding Kit (SQK-PBK004). In order to have compatible adapters for the addition of ONT barcodes the RP and FP primers used for the TeloPrime V2 Endpoint PCR need to be modified to include ONT primer binding sequences. 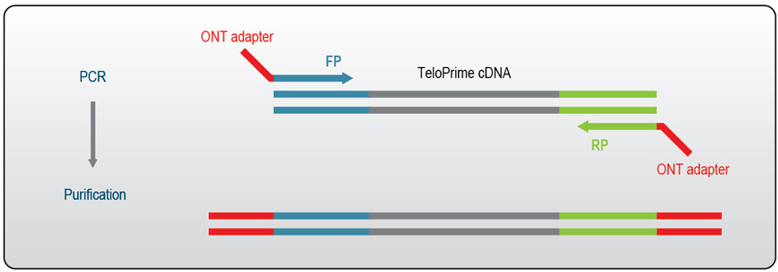
Figure 5 | Schematic for addition of ONT adapters to TeloPrime cDNA for barcoding and ONT sequencing.
The alternative FP and RP primer sequences required are listed below. These can be ordered as desalted oligos and should be used for the Endpoint PCR at a final concentration of 2 uM.
FP-ONT: 5′– TTTCTGTTGGTGCTGATATTGC TGGATTGATATGTAATACGACTCACTATAG –3'
RP-ONT: 5′– ACTTGCCTGTCGCTCTATCTTC TCTCAGGCGTTTTTTTTTTTTTTTTTT — 3'
*bold sequence is the standard FP and RP sequence and the 5' sequence is the ONT primer binding sequence.
The Endpoint PCR program should include one cycle at 50 degrees annealing temperature, then using 62 degrees annealing temperature for the remaining cycles.
