How Does TeloPrime V2 perform for different types of RNA?
The TeloPrime V2 PCR protocol has been validated with various RNA sources, including plants (Figure 1), different vertebrate species, such as sheep (Figure 2), and Universal Human Reference RNA (UHRR, Figure 3). The length distribution of the cDNA may vary according to the input RNA type. Note that the samples in Figures 1-3 have been run using different types of Bioanalyzer 2100 DNA Chip Assays.
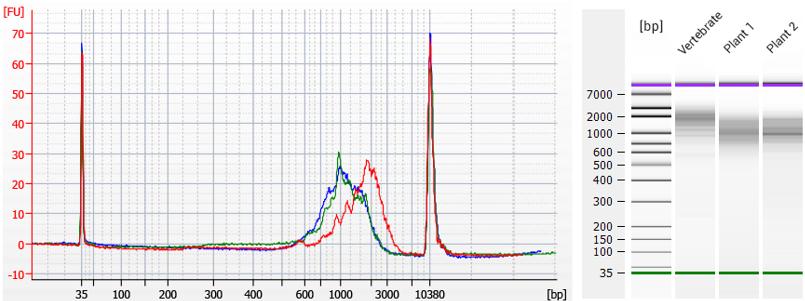
Figure 1 | TeloPrime cDNA prepared with V2 PCR Protocol for vertebrate and plant RNA. TeloPrime cDNA was generated from 1 µg of total RNA using a custom reverse transcription primer. The cDNA was amplified using a custom PCR reverse primer and the standard Forward PCR primer (FP), with 17 cycles (vertebrate, red trace) or 21 cycles (plant 1, blue trace, and plant 2, green trace) for endpoint PCR. PCR products were purified using 1x AMPure® PB beads (Pacific Biosciences) and normalized to a concentration of 300 pg/ul for analysis on a High Sensitivity DNA Chip (Bioanalyzer, Agilent Technologies). Customer-provided data reproduced with permission.
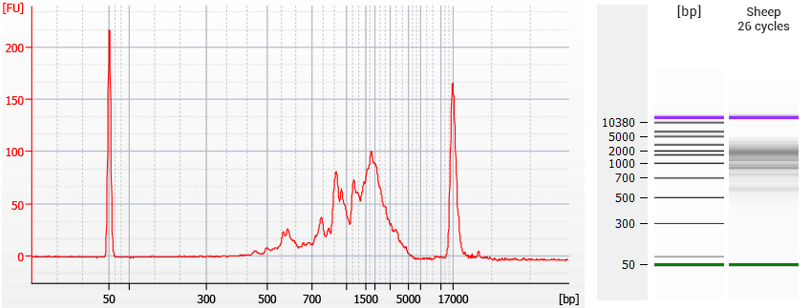
Figure 2 | TeloPrime cDNA prepared with V2 PCR protocol from sheep total RNA (RIN 8.4). TeloPrime cDNA was generated from 1 µg of total RNA, with standard TeloPrime RT primer and amplified with the standard TeloPrime V2 PCR primers. For endpoint PCR, 1 µl of cDNA was amplified with 26 cycles. The PCR product was purified using TeloPrime purification solutions and columns, and analyzed on a DNA 12000 Chip (Bioanalyzer, Agilent Technologies). Customer-provided data reproduced with permission.
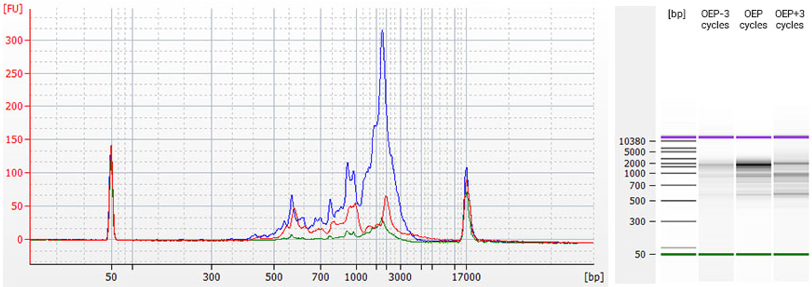
Figure 3 | TeloPrime cDNA generated with different PCR cycles. Endpoint PCR was performed using 2 µl of pooled TeloPrime cDNA with Optimal Endpoint PCR (OEP) cycle number (as calculated from qPCR, i.e., 80 % of maximum fluorescence), or using OEP ± 3 cycles. The amplified cDNA was analyzed on a Bioanalyzer DNA 12000 Chip (Agilent Technologies). Undercycling (green, OEP-3) produces low yields, while overcycling by 3 cycles (red, OEP +3) results in enrichment of shorter versus longer products and also reduces the yield. The optimal cycle number (blue, OEP) gives highest yield for longer cDNAs.
