What are the input requirements for QuantSeq FFPE?
The recommended input range is 10 - 200 ng total RNA from a FFPE sample. As a starting point, we recommend using >10 ng total RNA. Input amounts of >50 ng may be beneficial to detect low abundant transcripts efficiently. Lower input amounts (<10 ng) can also work but may lead to higher linker-linker artifact contamination, especially when the RNA quality is very low.
No prior poly(A) enrichment and/or rRNA depletion are required. DNase treatment is recommended (please see Should I perform a DNase treatment before QuantSeq FFPE?).
Below you can find an example of libraries generated from different input amounts of mouse liver FFPE RNA:
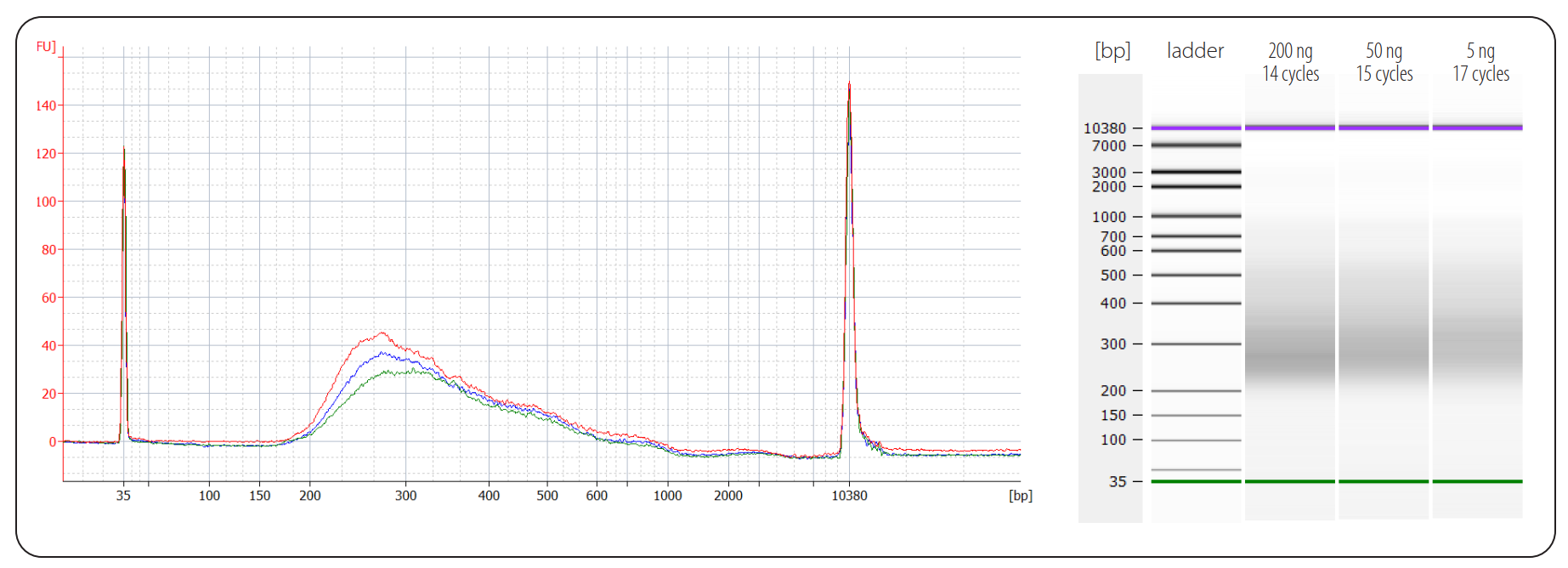
Figure | Bioanalyzer traces of QuantSeq FFPE libraries prepared from different input amounts of mouse liver FFPE RNA input. Libraries were prepared using 200 ng (red trace, 14 PCR cycles, 5 nM yield), 50 ng (blue trace, 15 PCR cycles, 4.9 nM yield) and 5 ng (green trace, 17 PCR cycles, 3.3 nM yield). Mouse liver FFPE RNA had a DV200 of 72 % (RIN 1.2). Endpoint PCR was performed using Lexogen’s 12 nt Unique Dual Indices.
