What is the typical library size distribution for CORALL FFPE libraries?
The library shape, size, and yield may vary depending on the sample input and quality. Heavily degraded FFPE samples typically generate shorter libraries than FFPE samples of higher quality.
Below are examples of CORALL FFPE libraries obtained from various input amounts of mouse liver FFPE RNA:

Figure | Fragment analyzer traces of typical CORALL FFPE libraries synthesized from Mus musculus (Mm) liver FFPE RNA (DV200 73 %, RQN 1.2). 200 ng (red trace), 50 ng (blue trace) and 5 ng (black trace) FFPE RNA treated with the DNA Removal Add-on (Cat. No. 235), then inserted into RiboCop rRNA depletion and a total RNA equivalent of 5 ng was used for CORALL FFPE library preparation. Libraries were amplified with 14 PCR cycles.
And CORALL FFPE libraries obtained from various sample types and input amounts:
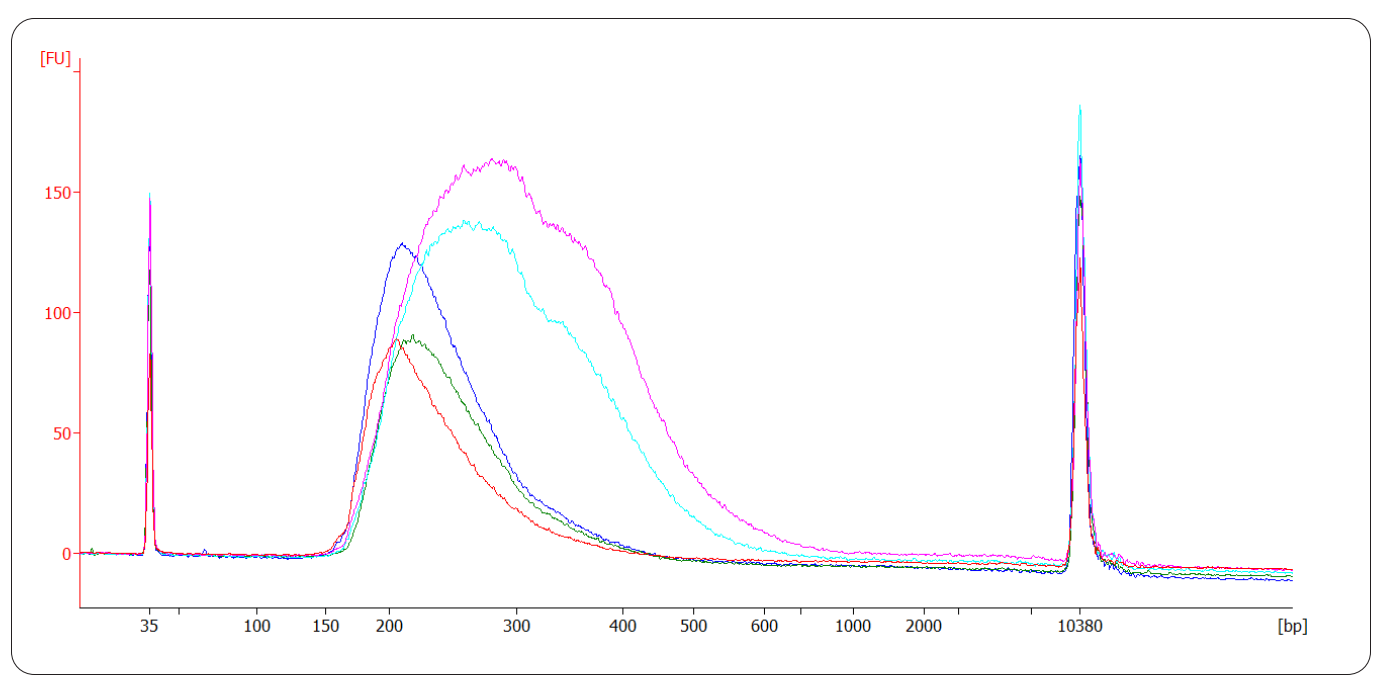
Figure | Bioanalyzer trace of ribo-depleted FFPE RNA of different quality inserted into CORALL FFPE library preparation. Input amounts listed refer to the total FFPE RNA inserted into DNA removal and RiboCop before the output is channeled into CORALL. Red trace: 50 ng Mm spleen FFPE RNA, DV200: 9 %, 15 cycles, average library size 256 bp. Blue trace: 20 ng human kidney tumor FFPE RNA, DV200: 45 % 13 cycles, average library size 260 bp. Green trace: 20 ng human kidney normal FFPE RNA, DV200: 50 % 12 cycles, average library size 273 bp. Turquoise trace: 10 ng mouse liver FFPE RNA, DV200: 79 % 12 cycles, average library size 327 bp. Pink trace: 10 ng mouse liver fresh frozen tissue RNA, RIN 9.1, 13 cycles, average library size 330 bp.
