Why do I see a peak at ~ 200 bp in my libraries?
What is this peak?
The presence of a peak at ~200 bp in QuantSeq FWD libraries is an indication of mis-hybridization.
Mis-hybridization in this case (the mis-priming of the oligo(dT) primers) can happen as a result of deviation from optimal incubation temperatures during first strand cDNA synthesis (i.e., reverse transcription).
How do I prevent mis-hybridization?
To prevent mis-hybridization, it is particularly important that during first strand cDNA synthesis, the RNA / FS1 reaction mix is kept at 42 °C after the denaturation step (steps 2-4 of the User Guide detailed protocol). Additionally, the FS2 / E1 mastermix should also be pre-warmed and kept at this same temperature (42°C) until is added to the samples (step 3).
NOTE: When using low input/degraded samples, step 2 is skipped and therefore, the FS1 / FS2 / E1 mastermix should be pre-warmed at 42°C before being added to the RNA samples (step 3). In addition, we recommend pre-heating the thermocycler block at 42°C before the samples are transferred on it (step 4).
ATTENTION: The centrifugation steps should always be carried out at room temperature (never at 4°C)!
TIP: Raising the reaction temperature to 50 °C for reverse transcription could also help prevent mis-priming. However, this can also result in a reduced yield overall and the qPCR assay should be used to ensure the libraries are amplified to provide sufficient yields for sequencing.
What does a mis-hybridization peak typically look like?
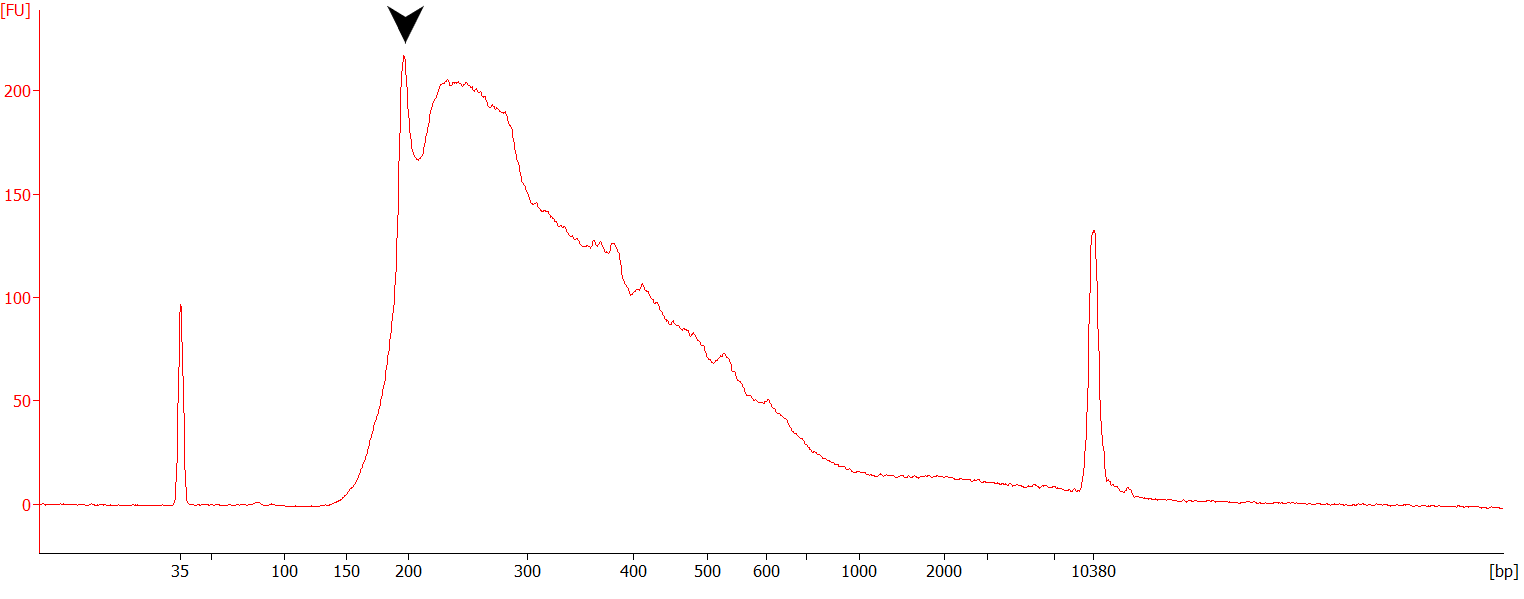
Figure |Example of a QuantSeq-FWD library prepared from 50ng of Universal Human Reference RNA (UHRR) and showing signs of mis-hybridization (peak at ~200 bp, black arrow).
What impact does mis-hybridization have on my results?
Mis-hybridization products correlate with a higher proportion of intronic and ribosomal RNA (rRNA) reads in the sequencing output. This happens because rRNAs contain A-rich sequence regions that oligo(dT) primers can more readily hybridize under less stringent conditions. As these non-specific inserts are small in size, these are efficiently amplified, hence, the presence of distinct peaks and elevated non-specific read fractions in your final libraries.
Other A-rich regions (present in various RNA molecules, including mRNAs) could also be captured and amplified in a similar way.
It is not possible to estimate the proportion of mis-hybridization products present in a library, before sequencing it.
The increased reads mapping to rRNAs and other mis-primed regions will reduce the proportion of reads of interest in the sequencing output. To compensate for this, when sequencing libraries with mis-hybridization peaks the sequencing depth can be increased (e.g., 5 - 10 M reads per sample).
Should I sequence libraries with mis-hybridization peaks?
Technically speaking, libraries with mis-hybridization peaks can be sequenced. However, it is important to note that the presence of non-specific hybridization does mean your data will include more non-specific “noise” than you would see in a library that was generated without mis-hybridization (as explained above).
To ensure maximum data quality, libraries should best be re-prepped, taking care of handling temperatures to avoid mis-hybridization effects, where possible.
End users must evaluate the risks and benefits of continuing with sequencing, vs. re-prepping the libraries, according to their own needs. The followings are provided as guidelines only to assist with deciding whether or not to rep-rep or sequence libraries with mis-hybridization signatures:
Reasons to re-prep libraries with mis-hybridization:
Sufficient amounts of RNA samples are available.
The results must be compared with previous libraries, or other library preps that do not show evidence of mis-hybridization.
Sufficient time / kit reagents / budgets are available to facilitate re-prepping.
Data quality is more important than time / cost.
Additional sequencing depth cannot be accommodated.
Reasons to sequence libraries with mis-hybridization:
Precious samples that cannot be re-prepped.
All samples within the batch / project show similar mis-hybridization features.
Results are stand-alone and won’t be compared to other sample batches or projects.
Insufficient time / budget / reagents available to re-prep libraries.
Additional noise / deviations in data quality can be tolerated.
Additional sequencing depth can be accommodated.
NOTE: The decision of whether or not to sequence libraries with mishybridization signatures resides with the end user. Lexogen provides no guarantees or minimum data quality thresholds for libraries that display mis-hybridization peaks.
