How do I calculate endpoint cycle numbers?
These guidelines are relevant for calculating the correct number of cycles to use for endpoint PCR with the following kits: QuantSeq V2 with UDIs, QuantSeq-Pool, CORALL RNA-Seq V2 and LUTHOR HD. Please consult the respective User Guides for kit-specific details.
The final number of Endpoint PCR cycles to use is calculated by the following formula:
No. Endpoint PCR cycles = 50% MF cycle No. – 3
50% MF cycle No. = cycle number corresponding to 50% of the maximum fluorescence value that is reached at the amplification plateau.
3 cycles are subtracted from the 50 % MF cycle number because the input used for Endpoint PCR (17 μl of cDNA) is 10X the input used for the qPCR assay (1.7 μl of cDNA). This corresponds to a difference in ~ 3 PCR cycles.
NOTE: It is usually safe to add one additional cycle to the calculated No. Endpoint PCR cycles without risking over-cycling.
Tips for calculating Endpoint PCR cycle numbers correctly:
Set the y-axis of the amplification plot to linear scale.
Ensure ROX background fluorescence is turned off.
Draw a horizontal line from the amplification plateau to the y-axis and record the value as the Maximum fluorescence value.
Calculate half of this value to get the 50 % MF value.
Draw a horizontal line from this value on the y-axis to intersect the amplification curve, then draw a vertical line from the intersection point to the x-axis.
The cycle number that this vertical line intersects is the 50 % MF cycle No. Round up to the next whole number.
Subtract 3 cycles from this number to get the final Endpoint PCR cycle number.
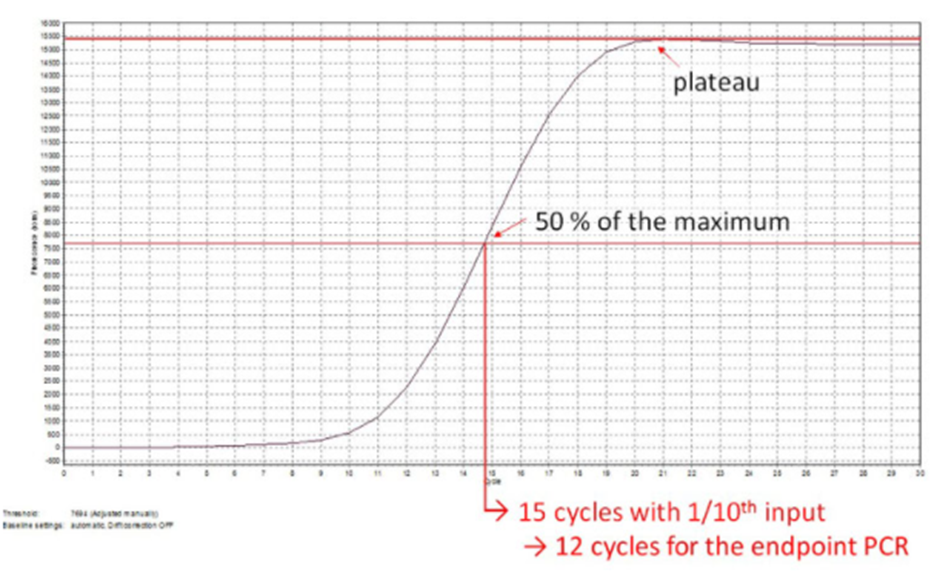
Figure | Example of Endpoint PCR cycle number calculation from qPCR amplification curve (linear scale).
